
The analysis is based on several librariesĬontaining ~9,000 biologically meaningful metabolite sets collected primarily from human studies Names with concentrations, or a concentration table. This module accepts a list of compound names, a list of compound MetaboAnalyst performs metabolite set enrichment analysis (MSEA) contains human and mammalian metabolite sets,Īs well as chemical class metabolite sets. The results can be explored in an interactive Upset diagram.
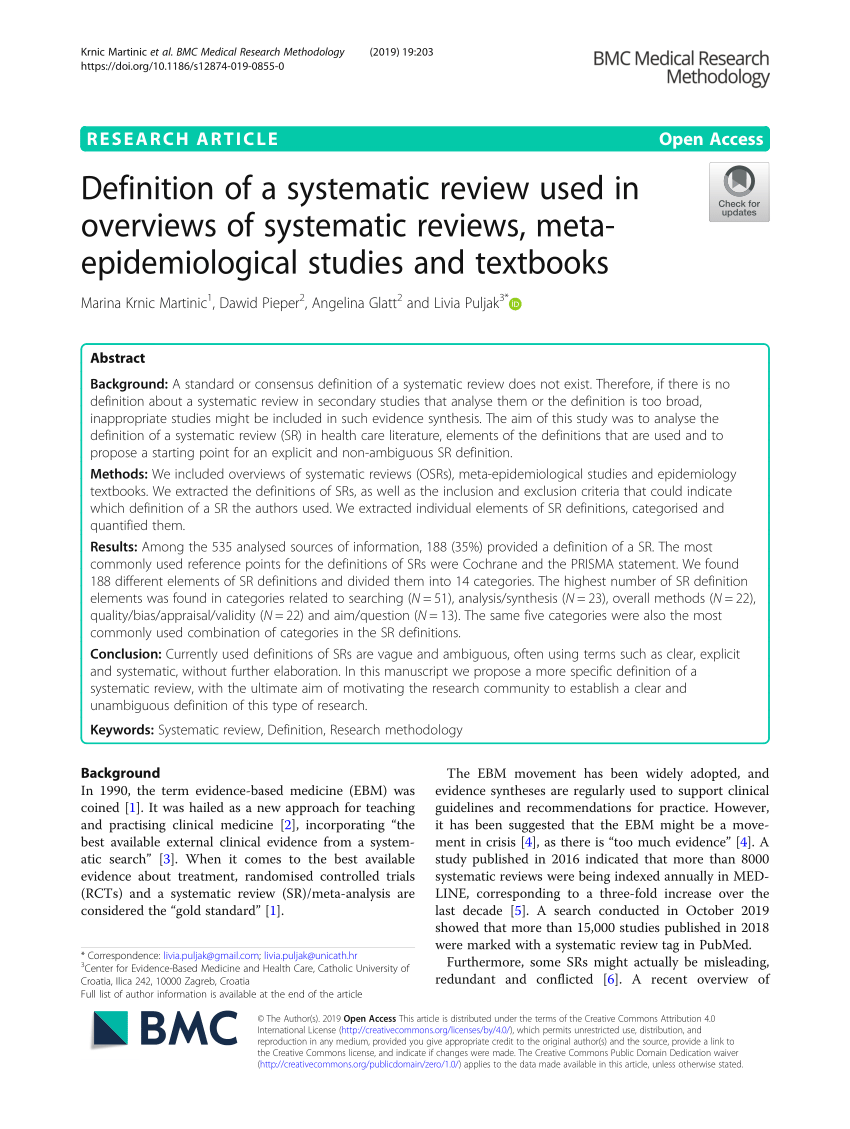
It currently supports several meta-analysis methods based on p-value combination, (compounds or annotated peaks) across multiple studies. Users can upload several annotated metabolomics data sets collected under comparable conditions to identify robust biomarkers In addition, users can manually select biomarkers or set up hold-out samples for flexible It offers classical univariate ROC curve analysis as well as more modern multivariate ROC curve analysisīased on PLS-DA, SVM or Random Forests. MetaboAnalyst provides the receiver operating characteristic (ROC) curve based approach for identifying potential biomarkersĪnd evaluating their performance. Including two-way ANOVA, multivariate empirical Bayes time-series analysis (MEBA), and ANOVA-simultaneous component analysis (ASCA). For two-factors / time-series data, users have more options Together with PCA and heatmaps for visual explorations.

It employs general linear models to accommodate modern epidemiological study, MetaboAnalyst now allows users to visualize and compute associations between phenotypes and metabolomics features with considerations of Orthogonal partial least squares-discriminant analysis (OPLS-DA) clustering - dendrogram, heatmap, K-means,Īnd self organizing map (SOM) as well as supervised classification - random forests and support vector machine (SVM).

Multivariate - principal component analysis (PCA), partial least squares-discriminant analysis (PLS-DA) and (and metabolites) (SAM) and empirical Bayesian analysis of microarrays (and metabolites) (EBAM) The objective is to enable high-throughput analysis for both targeted and untargeted metabolomics,Īnd to narrow the gap from raw spectra to biological insights.Ī wide array of commonly used statistical and machine learning methods are available: univariate - fold change, t-test, volcano plot,ĪNOVA, correlation analysis advanced feature selection - significance analysis of microarrays Normalization, statistical analysis, functional analysis, meta-analysis as well as integrative analysis with other The current MetaboAnalyst (V5.0) supports raw MS spectra processing, comprehensive data Over the past decade, MetaboAnalyst has evolved to become the most widely used platform (>300,000 users) in the metabolomicsĬommunity. MetaboAnalyst is a comprehensive platform dedicated for metabolomics data analysis via user-friendly, web-based interface.


 0 kommentar(er)
0 kommentar(er)
